nextPYP is an open-source, end-to-end platform for high-resolution single-particle cryo-EM/ET image analysis
cryo-EM/ET
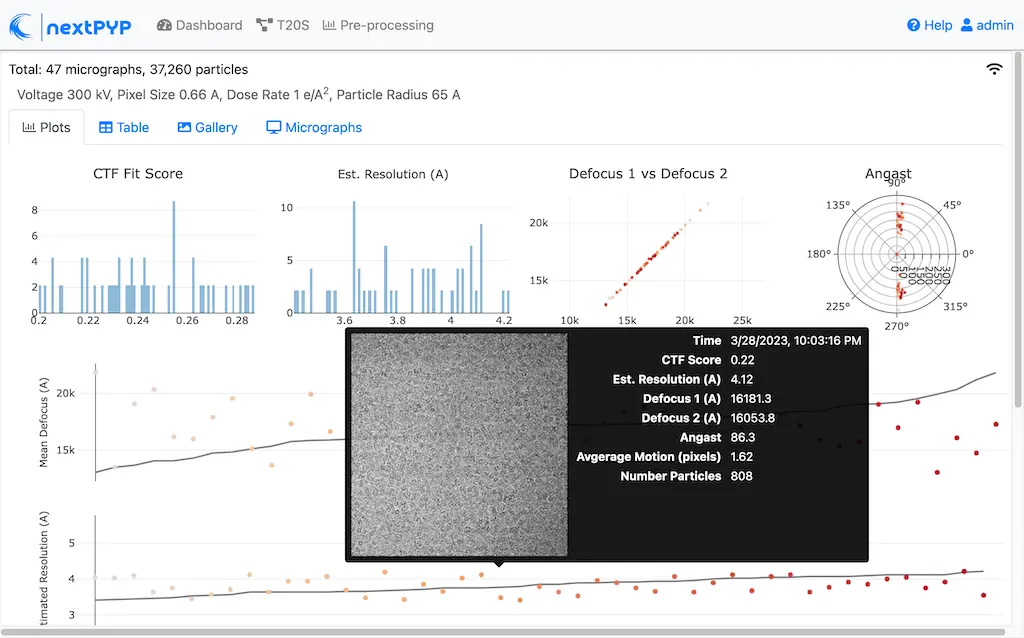
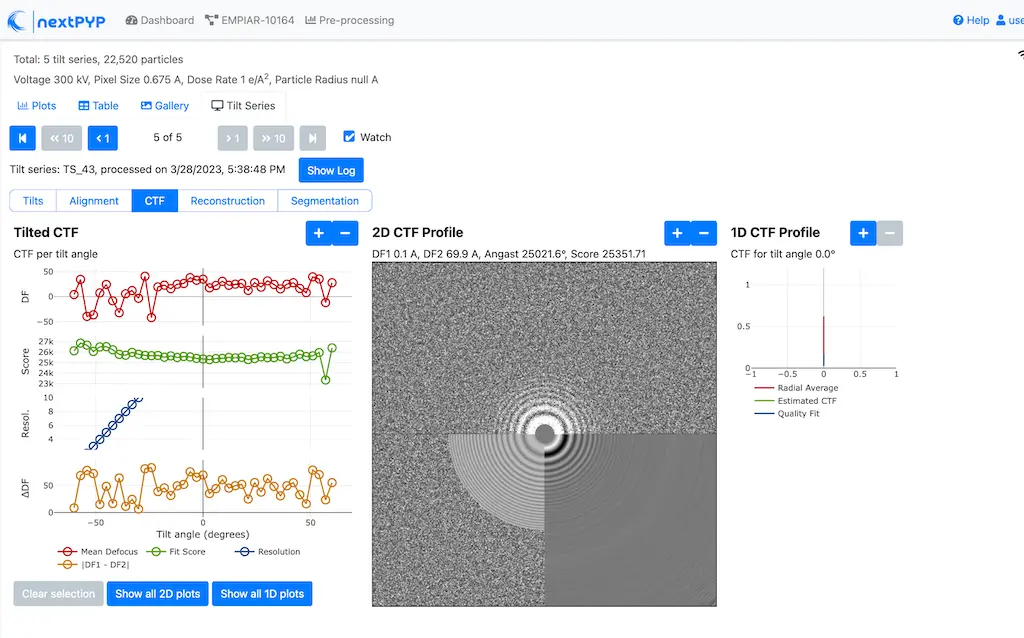
Analyze micrographs and tilt-series in real-time, including movie frame alignment, CTF estimation and particle picking. Learn more
2D/3D
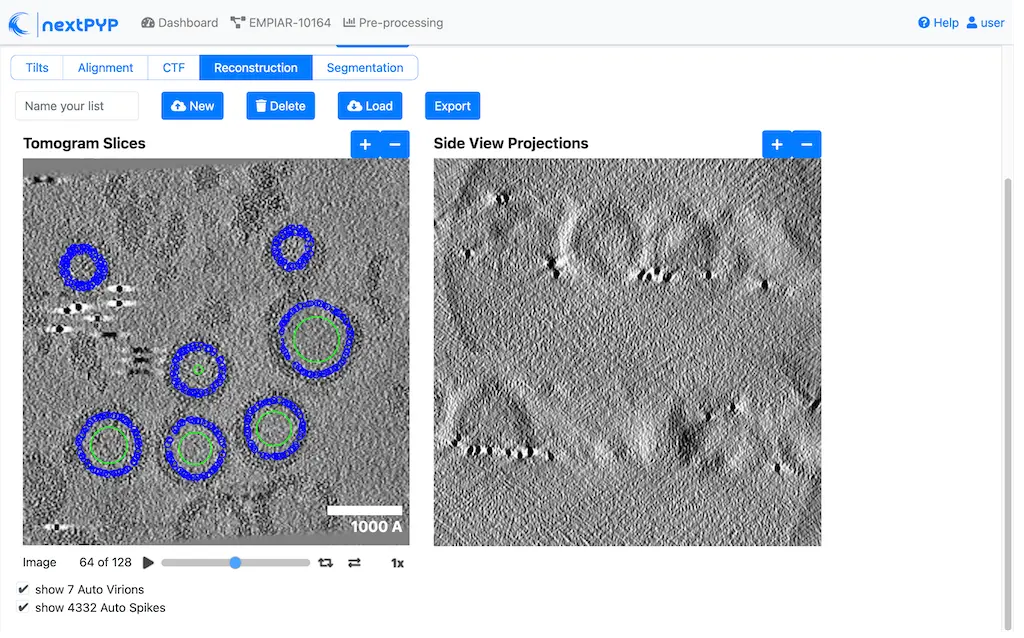
Pick particles from micrographs or tomograms using size-based, geometry-based, template-search, or neural network-based methods. Learn more
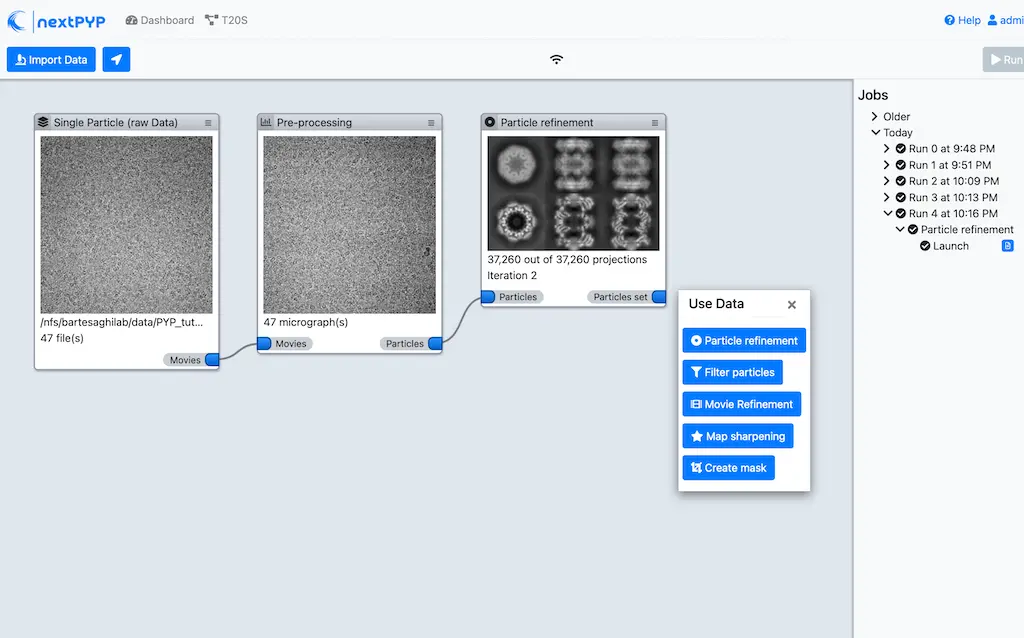
Single-particle/Sub-tomogram averaging
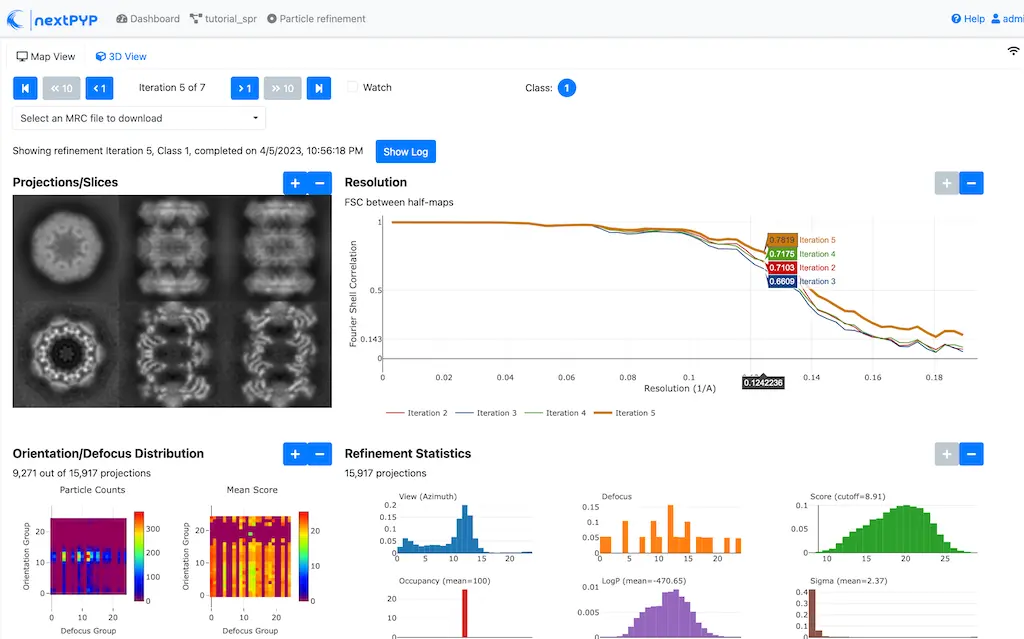
Multi-mode constrained refinement and classification. Particle-based CTF refinement, movie frame alignment and exposure weighting. Learn more
nextPYP features a storage-efficient architecture that avoids saving full-size tomograms, sub-volumes, and particle stacks, allowing routine processing of thousands of micrographs or tilt-series and tens of millions particle projections.
Try the live demo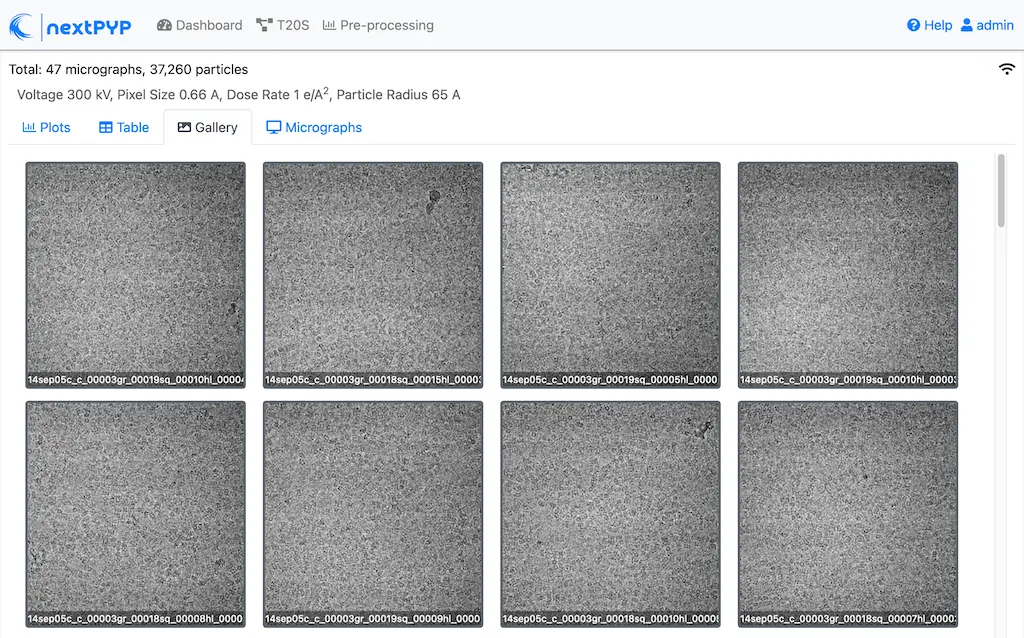
Watch the lectures and practical sessions from the nextPYP tutorial.
Course materials are also available.
Development of nextPYP is supported by grants from NIH/NIGMS (R01-GM141223), NIH/NIAID (U54-AI170752), the Chan Zuckerberg Initiative (2021-234602), and Duke University.
Liu, HF., Zhou, Y., Huang, Q., Martin, J., Bartesaghi, A., (2025) In situ structure determination of conformationally flexible targets using nextPYP, Nature Protocols, in press.
Huang, Q., Zhou, Y., Bartesaghi, A., (2024) MiLoPYP: self-supervised molecular pattern mining and particle localization in situ, Nature Methods, 21(10):1863-1872.
Liu, HF., Zhou, Y., Huang, Q., Piland, J., Jin, W., Mandel, J., Du, X., Martin, J., Bartesaghi, A., (2023) nextPYP: a comprehensive and scalable platform for characterizing protein variability in-situ using single-particle cryo-electron tomography, Nature Methods, 20(12):1909–1919.
Bouvette, J., Liu, HF., Du, X., Zhou, Y., Sikkema, AP., da Fonseca Rezende e Mello, J., …, Borgnia, MJ., Bartesaghi. A., (2021) Beam image-shift accelerated data acquisition for near-atomic resolution single-particle cryo-electron tomography, Nature Communications 12(1):1957.
Liu, HF., Zhou, Y., Bartesaghi, A. (2022) High-resolution structure determination using high-throughput electron cryo-tomography, Acta Crystallographica Section D: Structural Biology 78(7):817-824.
Huang, Q., Zhou, Y., Liu, HF., Bartesaghi, A. (2022) Accurate Detection of Proteins in Cryo-Electron Tomograms from Sparse Labels, European Conference on Computer Vision (ECCV) 13681:644-660.
Huang, Q., Zhou, Y., Liu, HF., Bartesaghi, A. (2022) Weakly Supervised Learning for Joint Image Denoising and Protein Localization in Cryo-Electron Microscopy, IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), 3246-3255.
Zhou, Y., Moscovich, A., Bendory, T., Bartesaghi, A., (2019) Unsupervised particle sorting for high-resolution single-particle cryo-EM, Inverse Problems 36:044002.
Bartesaghi, A., Aguerrebere, C., Falconieri, V., Banerjee, S., Earl, L. A., Zhu, X., …, Subramaniam, S. (2018). Atomic resolution cryo-EM structure of β-galactosidase, Structure 26(6):848-856.
Bartesaghi, A., Lecumberry, F., Sapiro, G., Subramaniam, S., (2012) Protein secondary structure determination by constrained single-particle cryo-electron tomography, Structure 20(12):2003-2013.
Jin W., Zhou Y., Bartesaghi, A. (2024). Accurate size-based protein localization from cryo-ET tomograms, Journal of Structural Biology: X, 10:100104.
Watson A., Bartesaghi, A. (2024) Advances in cryo-ET data processing: meeting the demands of visual proteomics, Current Opinion in Structural Biology, 87:102861.
Zhou Y., Moscovich A., Bartesaghi, A. (2022). Data-driven determination of number of discrete conformations in single-particle cryo-EM, Computer Methods and Programs in Biomedicine, 221:106892.