nextPYP is an open-source, end-to-end platform for high-resolution single-particle cryo-EM/ET image analysis
cryo-EM/ET
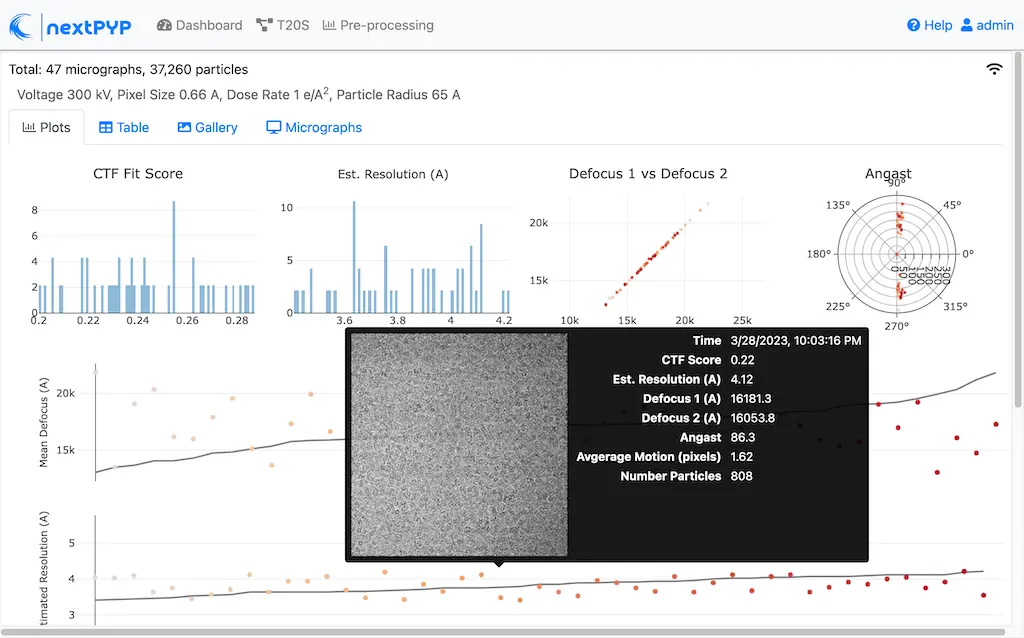
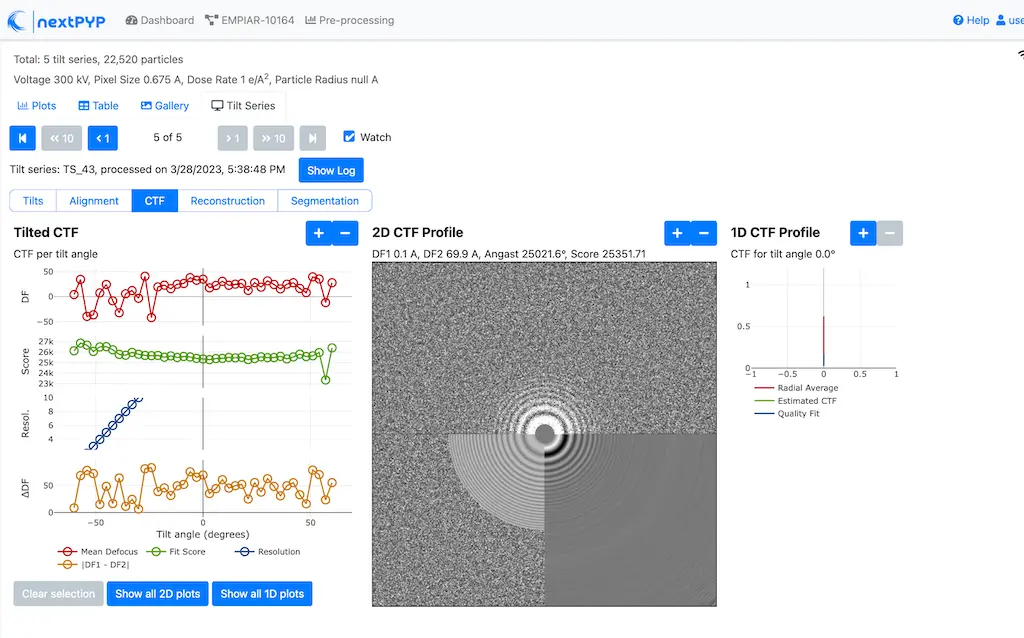
Analyze micrographs and tilt-series in real-time, including movie frame alignment, CTF estimation and particle picking. Learn more
2D/3D
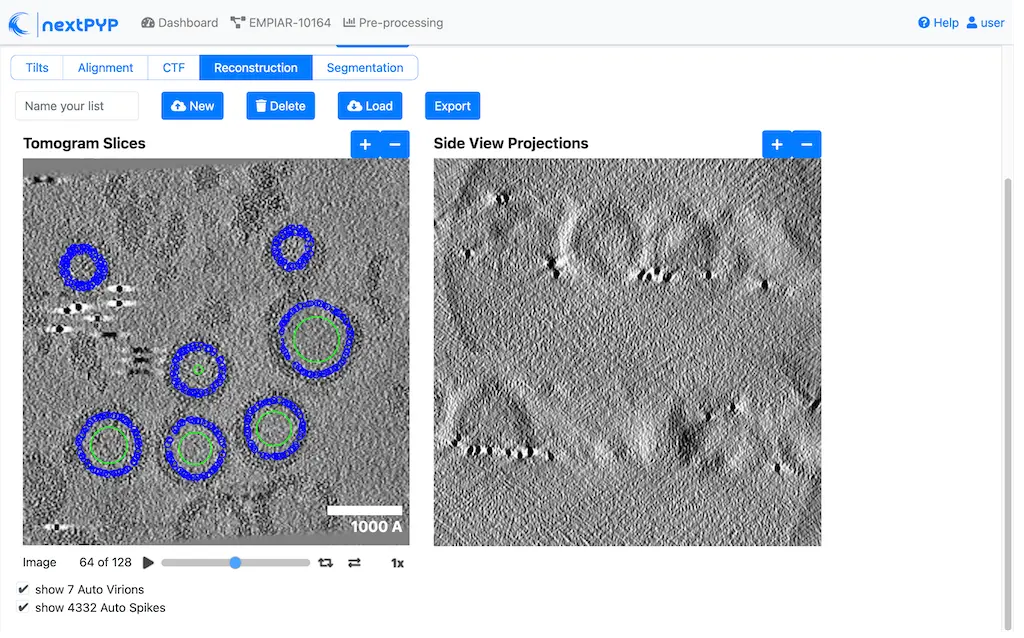
Pick particles from micrographs or tomograms using size-based, geometry-based or neural network-based methods. Learn more
Single-particle/Sub-tomogram averaging
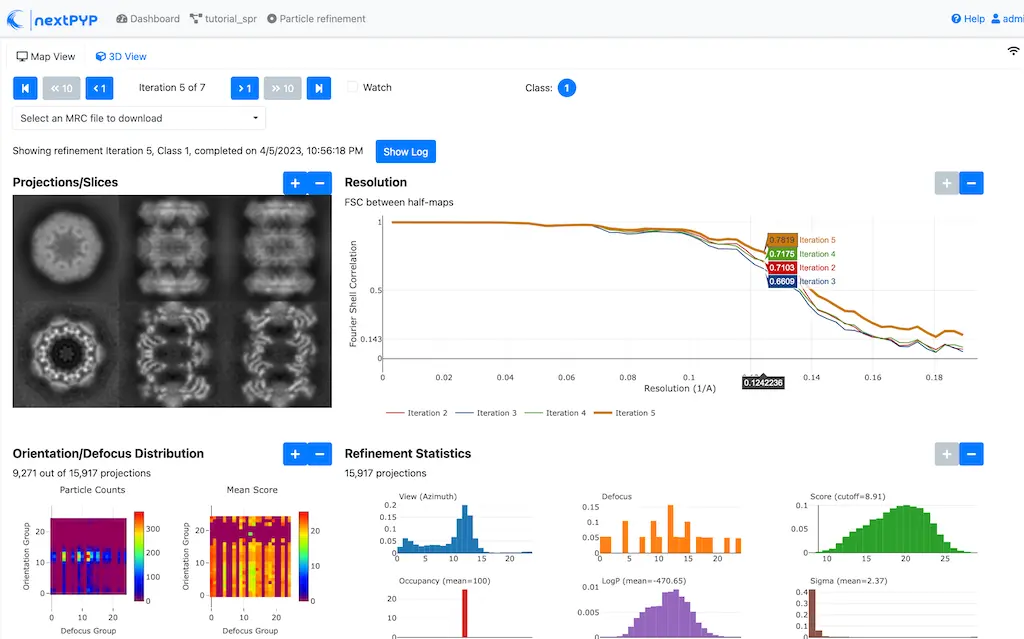
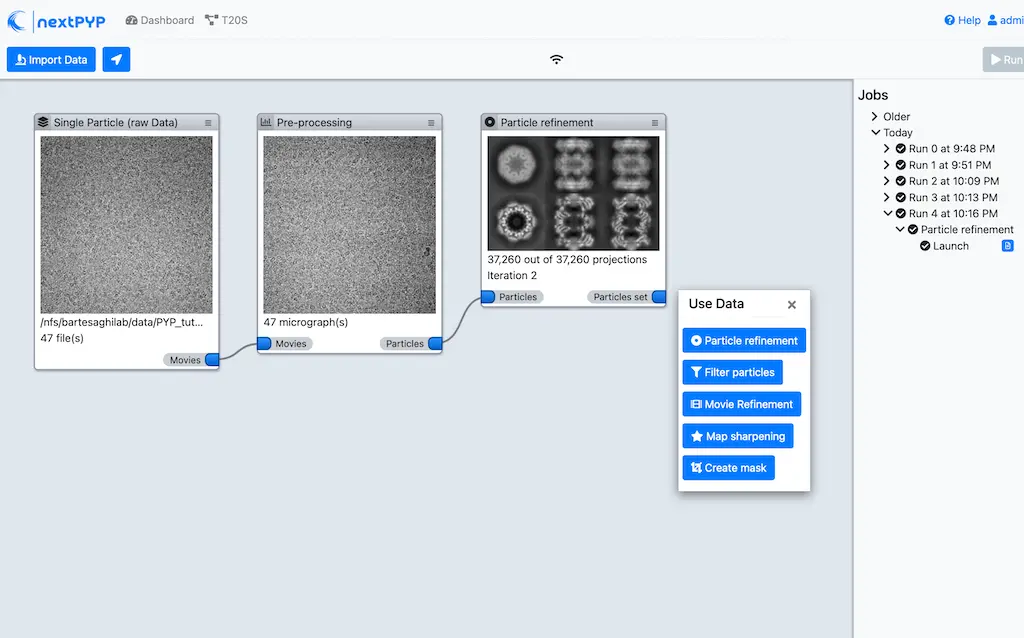
Multi-mode constrained refinement and classification. Particle-based CTF refinement, movie frame alignment and exposure weighting. Learn more
nextPYP features a storage-efficient architecture that avoids saving full-size tomograms, sub-volumes, and particle stacks, allowing routine processing of thousands of micrographs or tilt-series and tens of millions particle projections.
Try the live demo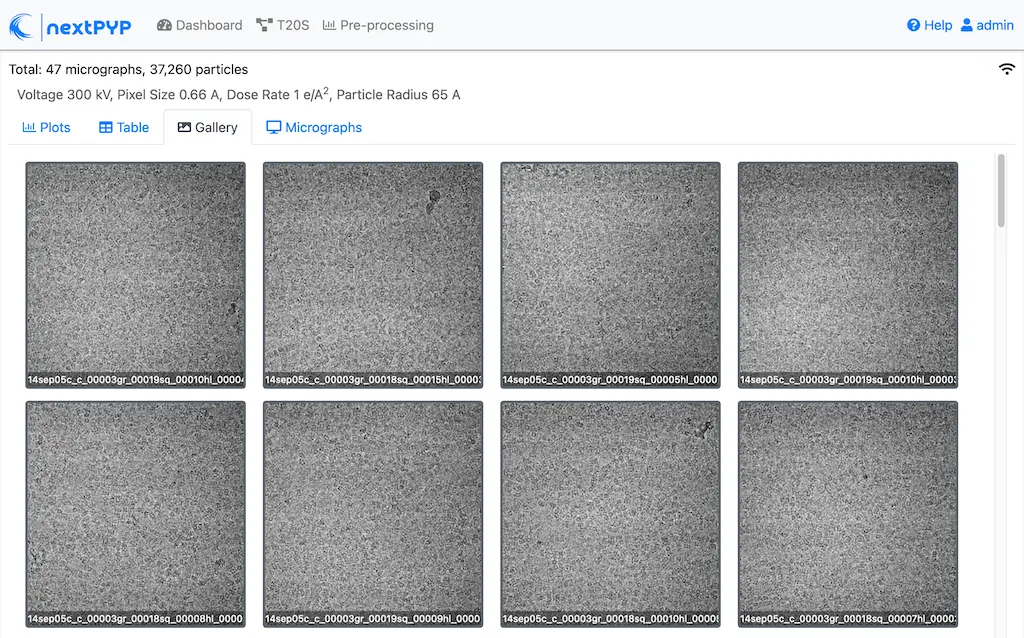
Watch the recorded lectures and practical sessions from the workshop.
Development of nextPYP is supported by grants from NIH/NIGMS (R01-GM141223), NIH/NIAID (U54-AI170752), the Chan Zuckerberg Initiative (2021-234602), and Duke University.
Liu, HF., Zhou, Y., Huang, Q., Piland, J., Jin, W., Mandel, J., Du, X., Martin, J., Bartesaghi, A., (2023) nextPYP: a comprehensive and scalable platform for characterizing protein variability in-situ using single-particle cryo-electron tomography, Nature Methods, 20, 1909–1919.
Bouvette, J., Liu, HF., Du, X., Zhou, Y., Sikkema, AP., da Fonseca Rezende e Mello, J., …, Borgnia, MJ., Bartesaghi. A., (2021) Beam image-shift accelerated data acquisition for near-atomic resolution single-particle cryo-electron tomography, Nature Communications 12, 1957.
Liu, HF., Zhou, Y., Bartesaghi, A. (2022) High-resolution structure determination using high-throughput electron cryo-tomography, Acta Crystallographica Section D: Structural Biology 78 (7), pp. 817-824.
Huang, Q., Zhou, Y., Liu, HF., Bartesaghi, A. (2022) Accurate Detection of Proteins in Cryo-Electron Tomograms from Sparse Labels, European Conference on Computer Vision (ECCV) 13681, pp. 644-660.
Huang, Q., Zhou, Y., Liu, HF., Bartesaghi, A. (2022) Weakly Supervised Learning for Joint Image Denoising and Protein Localization in Cryo-Electron Microscopy, IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), pp. 3246-3255.
Zhou, Y., Moscovich, A., Bendory, T., Bartesaghi, A., (2019) Unsupervised particle sorting for high-resolution single-particle cryo-EM, Inverse Problems 36, 044002.
Bartesaghi, A., Aguerrebere, C., Falconieri, V., Banerjee, S., Earl, L. A., Zhu, X., …, Subramaniam, S. (2018). Atomic resolution cryo-EM structure of β-galactosidase, Structure 26(6), pp. 848-856.
Bartesaghi, A., Lecumberry, F., Sapiro, G., Subramaniam, S., (2012) Protein secondary structure determination by constrained single-particle cryo-electron tomography, Structure 20(12), pp. 2003-2013.